Abstract
Introduction: The impact of the early expression of sickle hemoglobin (HbS) on erythroid differentiation remains unclear. We hypothesized that HbS already plays a detrimental role by altering the lipidome/proteome during sickle cell disease (SCD) erythropoiesis. Provided that in-vivo studies do not allow the dynamical follow-through of homogenous populations, are biased by non-cell autonomous interactions, and that bone marrow (BM) harvesting in SCD individuals is difficult to justify, we aimed to compare whole-cell/membrane proteomes and lipidomes in an ex-vivo human model of SCD and healthy erythropoiesis, as well as in mature peripheral blood (PB) erythrocytes.
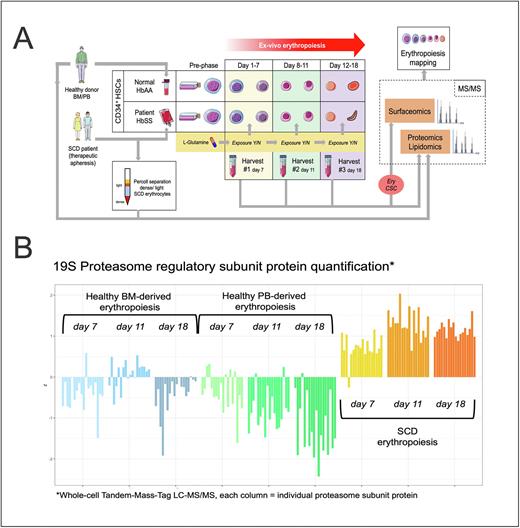
Methods: Healthy and SCD erythropoiesis was modeled ex-vivo in a 3-phase liquid system at ambient O2 tension starting with CD34+ hematopoietic stem cells (HSC) isolated either from healthy donor BM or GSCF-stimulated PB, and SCD patient apheresis products (Panel A). Whole cell protein extracts were obtained at 3 timepoints (day 7, 11, 18, i.e early-, late-erythroblast (EB) and reticulocyte stages). Membrane extracts were purified with a novel erythroid Cell-Surface-Capture method. Proteomes and lipidomes were characterized by quantitative LC-MS/MS and compared according to time-points and health vs. disease states in unbiased/unsupervised models. Multi-Omics Factor Analysis (MOFA) determined key modifiers of SCD erythropoiesis. Moreover, omic analyses were performed on mature healthy and SCD erythrocytes obtained from respective PB (after density-fractionation of light/dense cells for SCD). Proteasome subunits were quantified in healthy vs. SCD erythrocyte ghosts by WB. Proteasome activity was assayed in mature cells and in ex-vivo late-stage SCD erythroid cells under different L-glutamine levels.
Results: First, we observed that CD34+ HSCs derived from BM vs. PB generate significantly divergent proteomes/lipidomes in ex-vivo erythropoiesis. Specifically, differences in glutathione and glucose pathway proteins were observed. Amongst them, alpha-enolase is 2-fold to 6-fold depleted in intermediate and reticulocyte stages respectively, when comparing PB vs BM-derived erythropoiesis. This finding is relevant in consideration of prior data on the regulation of HSC erythroid vs myeloid lineage commitment by glucose/glutamine metabolism. Second, we used SCD vs BM-derived erythropoiesis (reference healthy control) to capture the most significant modifications. Among those, the proteasome was progressively abnormally localized in SCD. We observed the depletion of core 20S subunits and sequestration of 19S regulators at the membrane (Panel B), which could be confirmed in dense SCD erythrocyte membranes. Also, we observed an inversion of proteasome activity between membrane and cytosolic extracts in light vs. dense SCD erythrocytes (increased membrane activity in dense SCD erythrocytes). Removing glutamine from culture in the ex-vivo model of SCD erythropoiesis significantly increased the membrane proteasome activity at the reticulocyte stage. Moreover, oxidative stress defenses and cholesterol biosynthesis were perturbed in SCD erythropoiesis, including the upregulation of Heme-oxygenase-1. Also, the antioxidant protein balance was abnormal in mature-dense SCD erythrocyte membranes where insoluble aggregates (i.e. actin/ankyrin + HbS + GAPDH) were sequestered. Additionally, increased GAPDH could be seen in cytoskeleton extracts. Third, we observed the erythroid lipidome to evolve harmoniously in SCD vs healthy erythropoiesis across time-points, while significant perturbations were seen once erythrocytes are in circulation. In fact, dense SCD erythrocytes had significantly abnormal levels of lysophosphatidylcholines (depleted), sphingomyelins (depleted) and triacylglycerols (enriched). Fourth, MOFA revealed the key contribution of heme synthesis, glycolysis and proteasome regulation to the differences in global phenotype between SCD and healthy erythropoiesis.
Conclusion: Our study shows that a) ex-vivo human erythropoiesis models based on healthy PB vs BM-derived HSCs differ significantly on a lipidomic/proteomic level, highlighting the importance of source choice, b) HbS expression has an incremental proteomic impact during erythropoiesis and c) the lipidome is unaltered in SCD erythropoiesis but radically changes once erythrocytes are circulating.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal